InQuanto-NGLView
The InQuanto-NGLView extension provides basic utilities for visualizing chemical
systems via the NGLView package. These utilities return NGL widgets which
can be interactively viewed in a jupyter notebook. The functionality includes visualization of molecular structures,
molecular fragmentation schemes, and molecular orbital isosurfaces.
Visualizing Structures
The VisualizerNGL class is the central object of the InQuanto-NGLView extension.
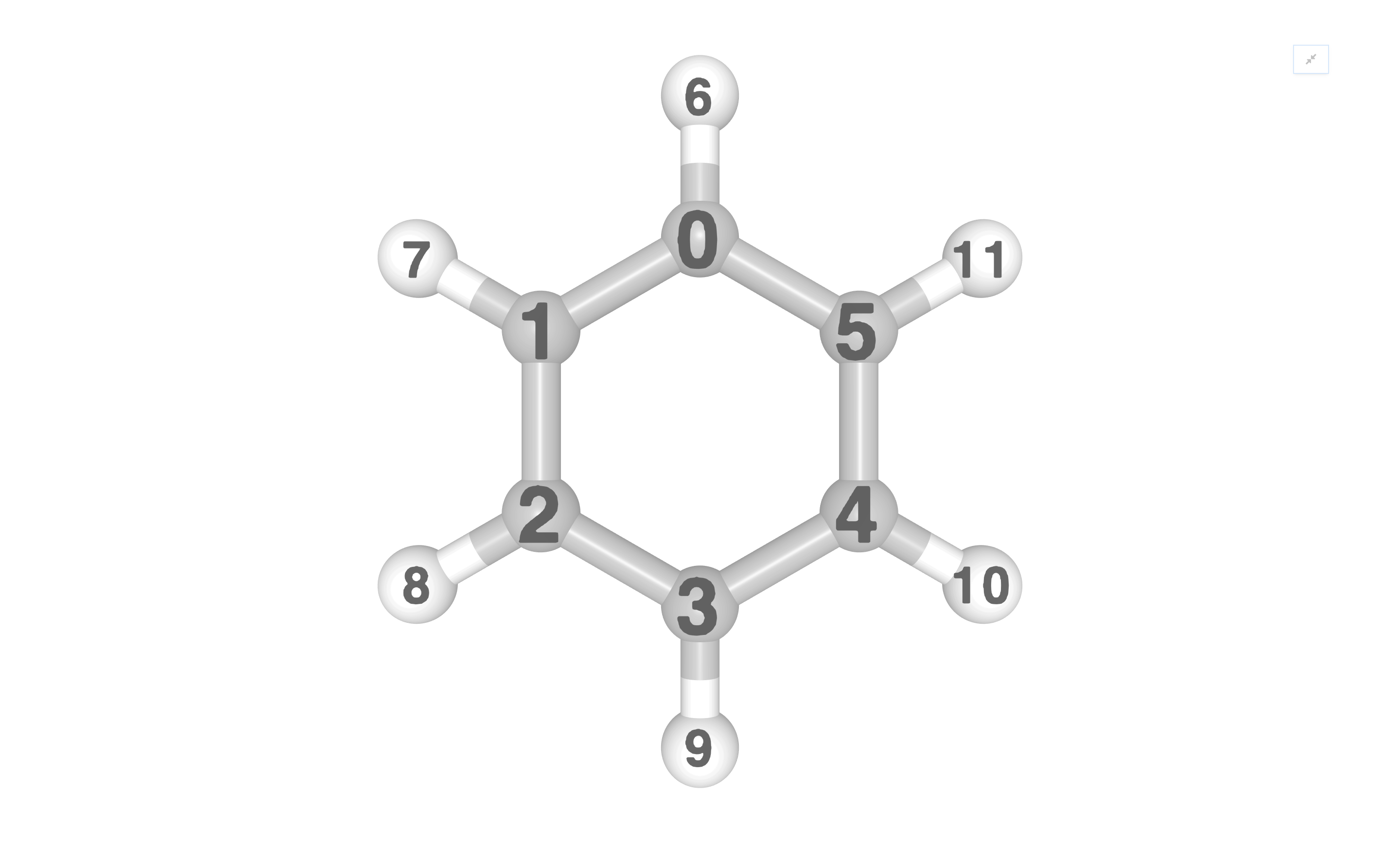
VisualizerNGL takes an InQuanto Geometry object as input,
and produces an interactive NGLWidget with the
visualize_molecule() method, visualizable in a jupyter notebook. See below
for an example with molecular geometry:
from inquanto.geometries import GeometryMolecular
from inquanto.extensions.nglview import VisualizerNGL
xyz = [
['C', [ 0.0000000, 1.4113170, 0.0000000]],
['C', [ 1.2222370, 0.7056590, 0.0000000]],
['C', [ 1.2222370, -0.7056590, 0.0000000]],
['C', [ 0.0000000, -1.4113170, 0.0000000]],
['C', [-1.2222370, -0.7056590, 0.0000000]],
['C', [-1.2222370, 0.7056590, 0.0000000]],
['H', [ 0.0000000, 2.5070120, 0.0000000]],
['H', [ 2.1711360, 1.2535060, 0.0000000]],
['H', [ 2.1711360, -1.2535060, 0.0000000]],
['H', [ 0.0000000, -2.5070120, 0.0000000]],
['H', [-2.1711360, -1.2535060, 0.0000000]],
['H', [-2.1711360, 1.2535060, 0.0000000]]
]
c6h6_geom = GeometryMolecular(xyz)
visualizer = VisualizerNGL(c6h6_geom)
visualizer.visualize_molecule(atom_labels="index")
Note
Code snippets on this page generate interactive cells in a jupyter notebook. Static images are shown here for demonstration.
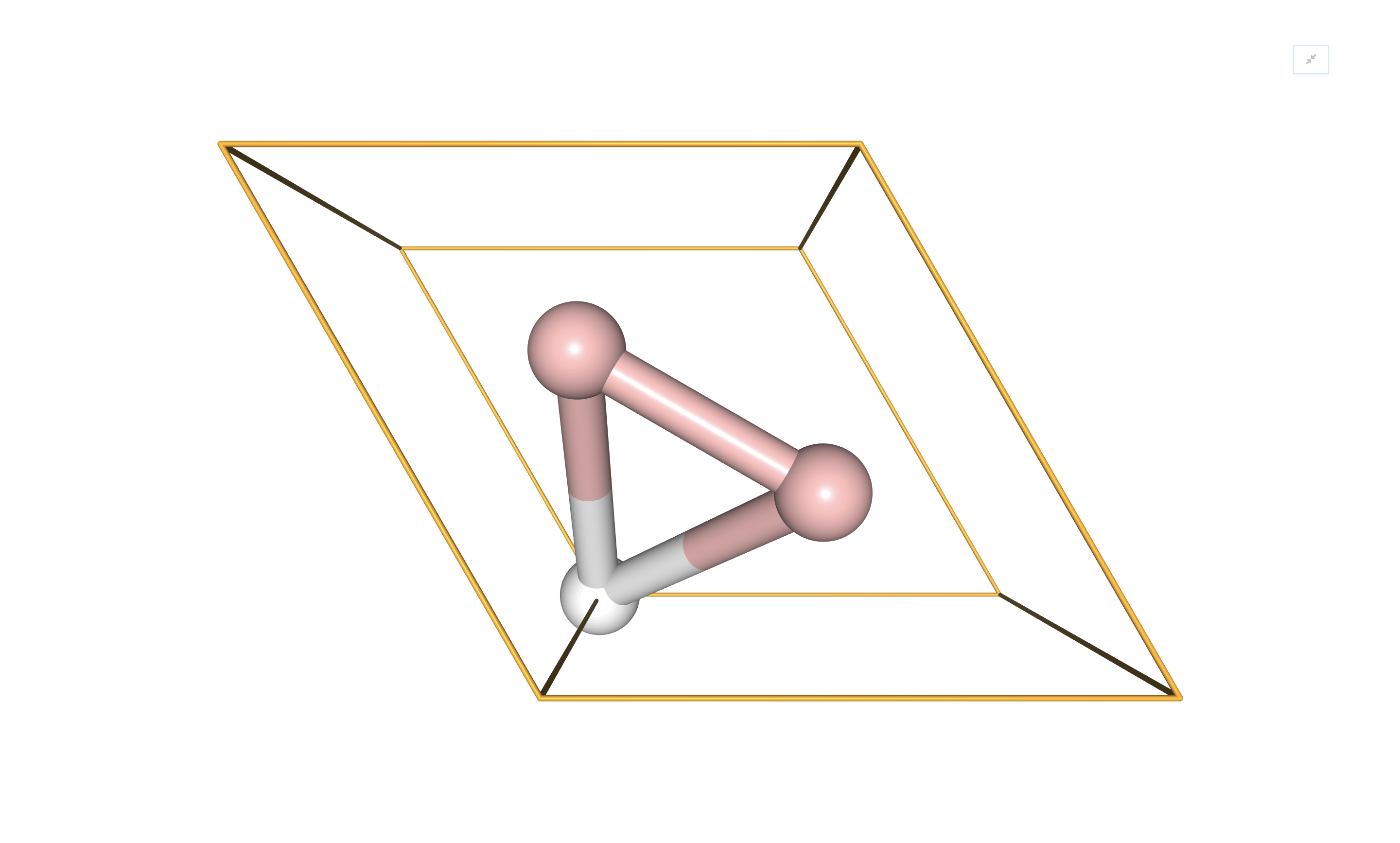
Similarly, periodic systems may be visualized by providing VisualizerNGL with a
GeometryPeriodic object:
from inquanto.extensions.nglview import VisualizerNGL
from inquanto.geometries import GeometryPeriodic
import numpy as np
# AlB2 unit cell
a=3.01 # lattice vectors in Angstroms
c=3.27
a, b, c = [
np.array([a, 0, 0]),
np.array([-a/2, a*np.sqrt(3)/2, 0]),
np.array([0, 0, c])
]
atoms = [
["Al", [0, 0, 0]],
["B", a/3 + b*2/3 + c/2],
["B", a*2/3 + b/3 + c/2]
]
alb2_geom = GeometryPeriodic(geometry=atoms, unit_cell=[a, b, c])
visualizer = VisualizerNGL(alb2_geom)
visualizer.visualize_unit_cell()
Visualizing Fragments
There are several fragmentation methods available in InQuanto; to visualize a fragmentation scheme defined in a
GeometryMolecular object, use the
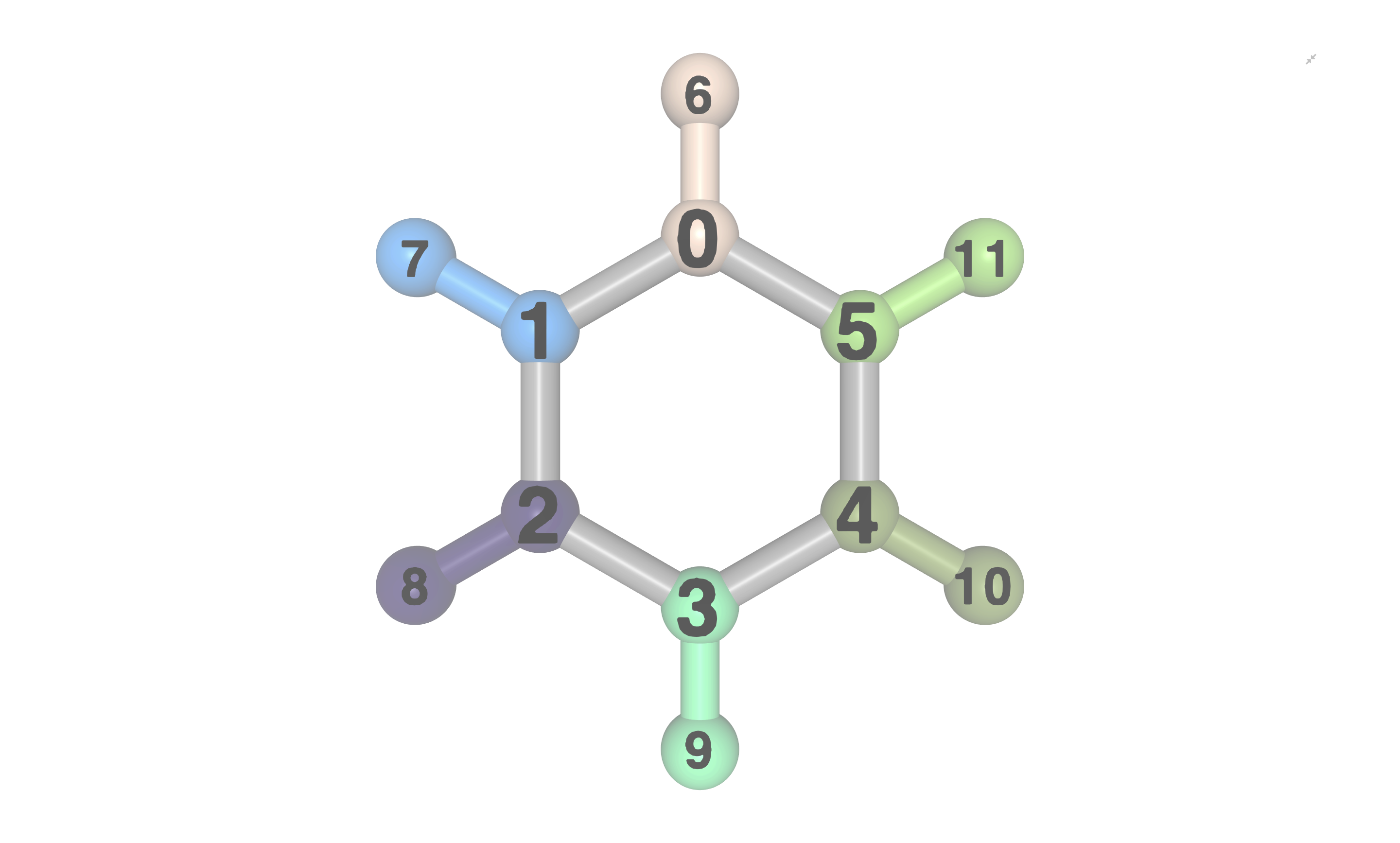
visualize_fragmentation() method:
c6h6_geom.set_groups(
"fragments",
{
"ch1": [0, 6],
"ch2": [5, 11],
"ch3": [4, 10],
"ch4": [3, 9],
"ch5": [2, 8],
"ch6": [1, 7]
}
)
visualizer.visualize_fragmentation("fragments", atom_labels="index")
Note
Visualizing fragments is not yet supported for periodic geometries.
Visualizing Orbitals
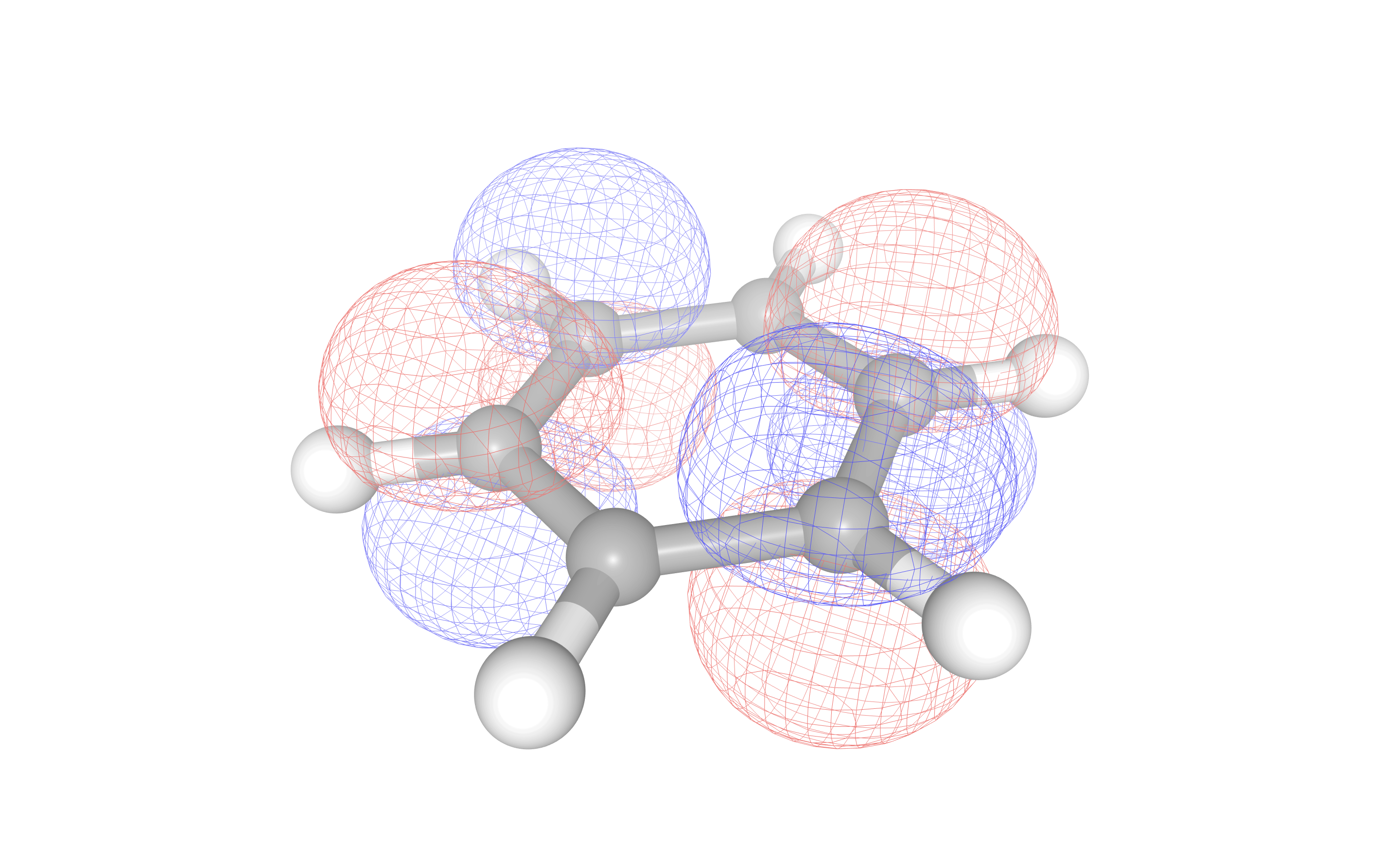
To provide a visual aid in active space selection, molecular orbitals may be visualized with the
visualize_orbitals() method. Orbital information must be provided in .cube
format, which may be generated for molecular systems with the InQuanto-pyscf extension. In the example
below, we generate the Hartree-Fock molecular orbitals for benzene in a minimal basis, and select the LUMO for
visualisation.
from inquanto.extensions.pyscf import ChemistryDriverPySCFMolecularRHF
driver = ChemistryDriverPySCFMolecularRHF(geometry=c6h6_geom.xyz, basis="sto3g")
cube_orbitals=driver.get_cube_orbitals()
ngl_mos = [visualizer.visualize_orbitals(orb) for orb in cube_orbitals]
ngl_mos[21]
Finally we note that extended NGLView options can be exposed by rendering its graphical user interface (GUI) using the following option:
g = GeometryMolecular(xyz)
visualizer = VisualizerNGL(g)
visualizer.visualize_molecule().display(gui=True)